The Effect of Spatial Organization of Chromatin on Morphology of
Eukaryotic Cell Nucleus
-NUCCHROM-


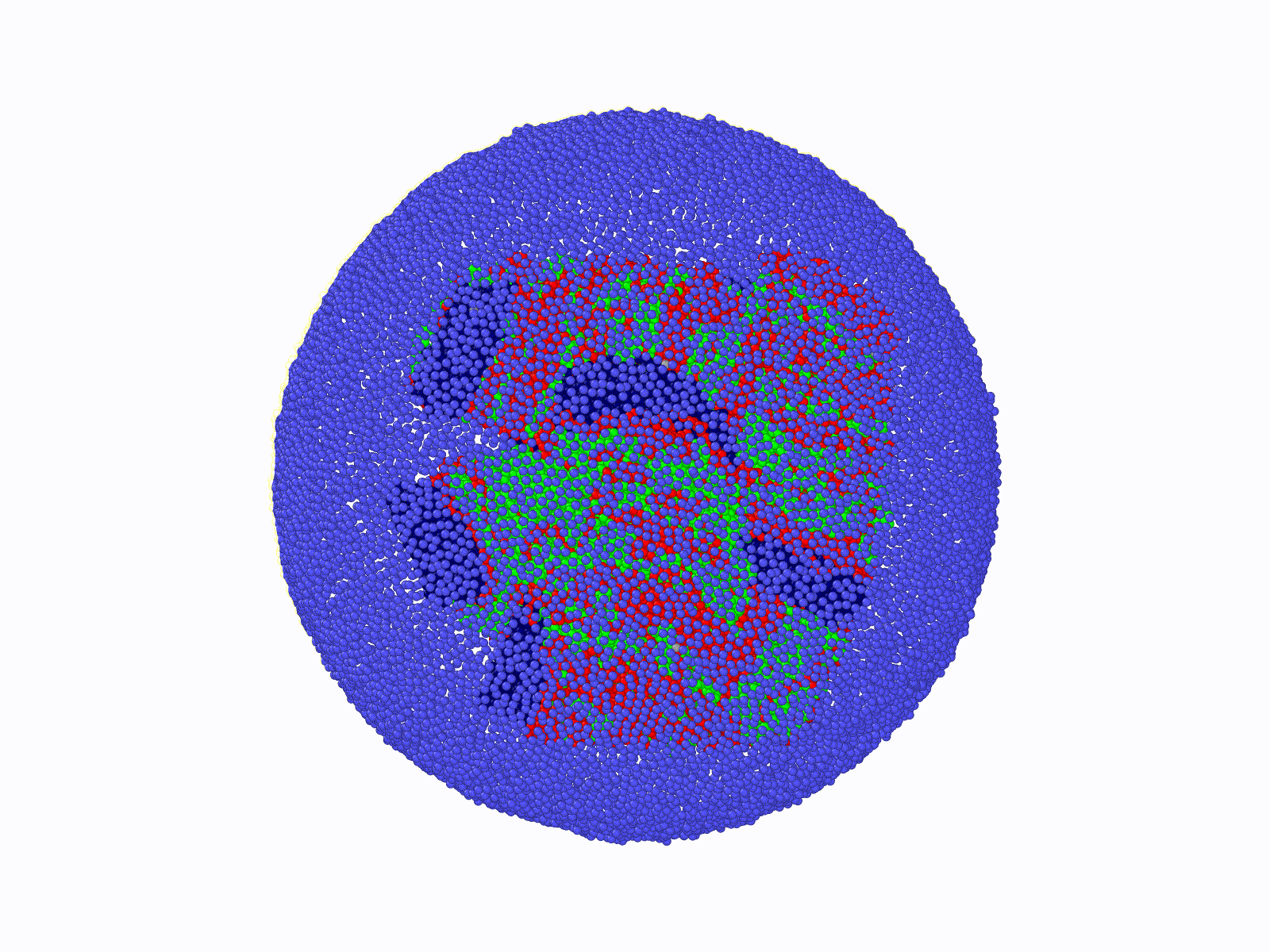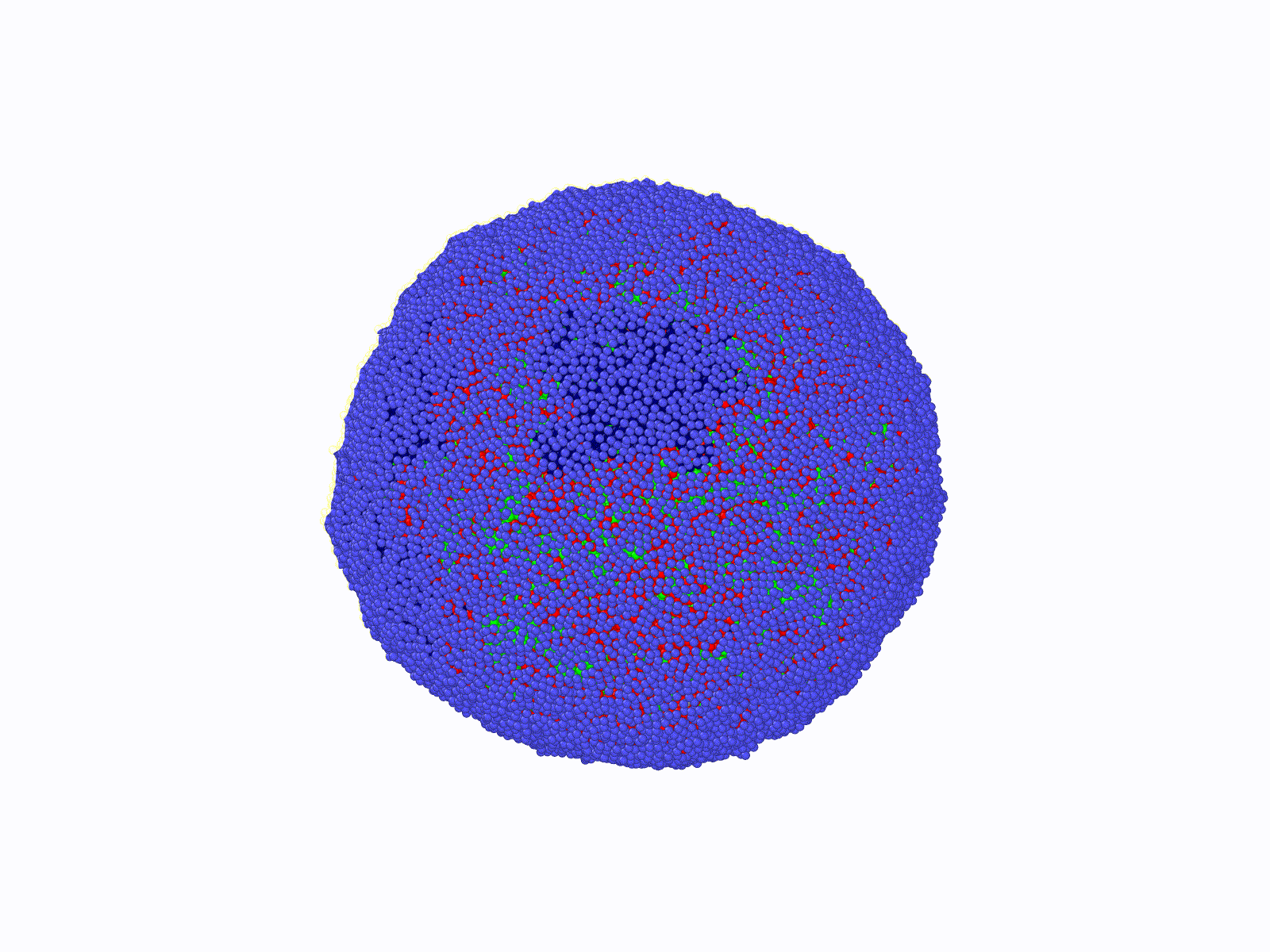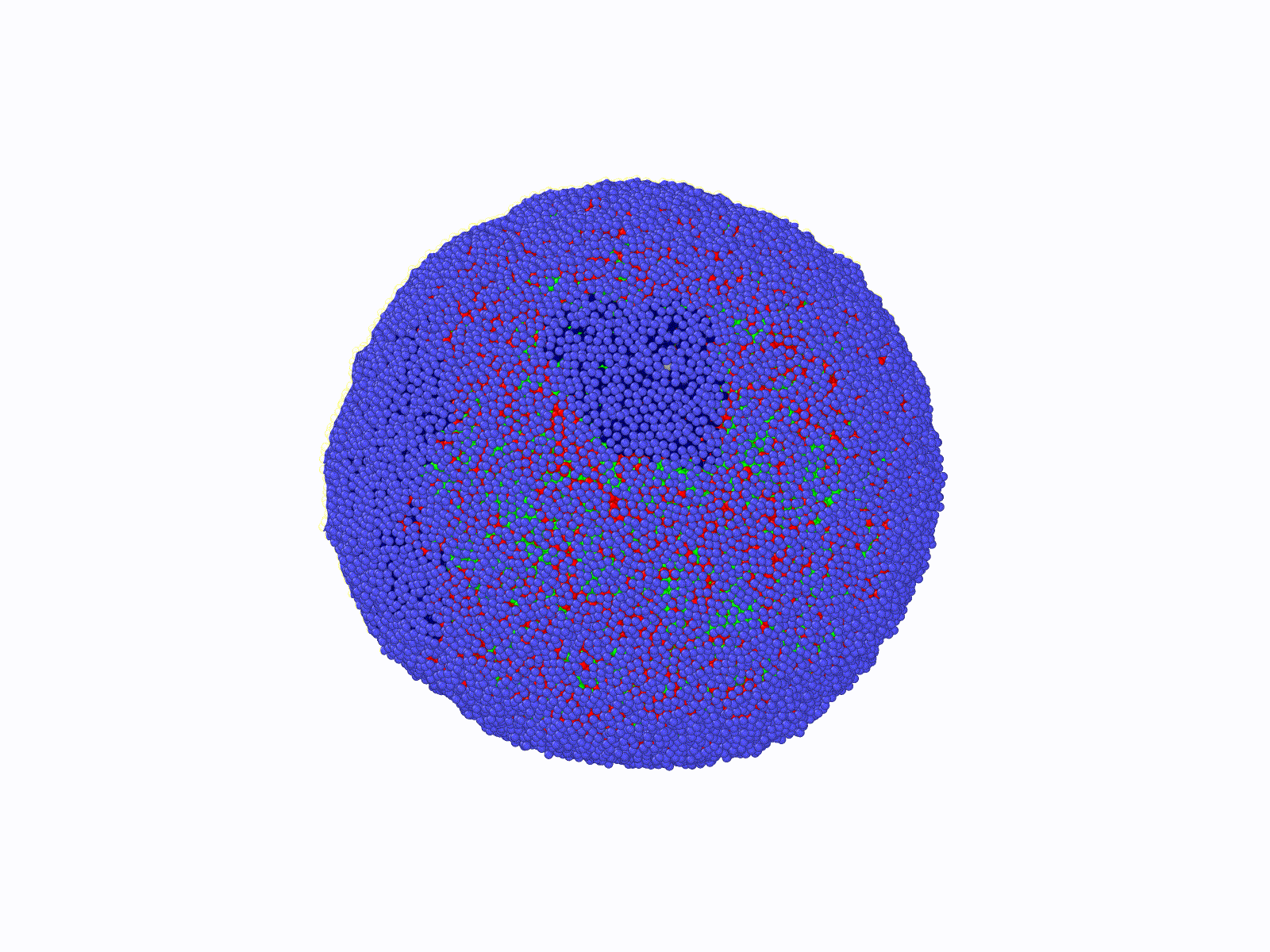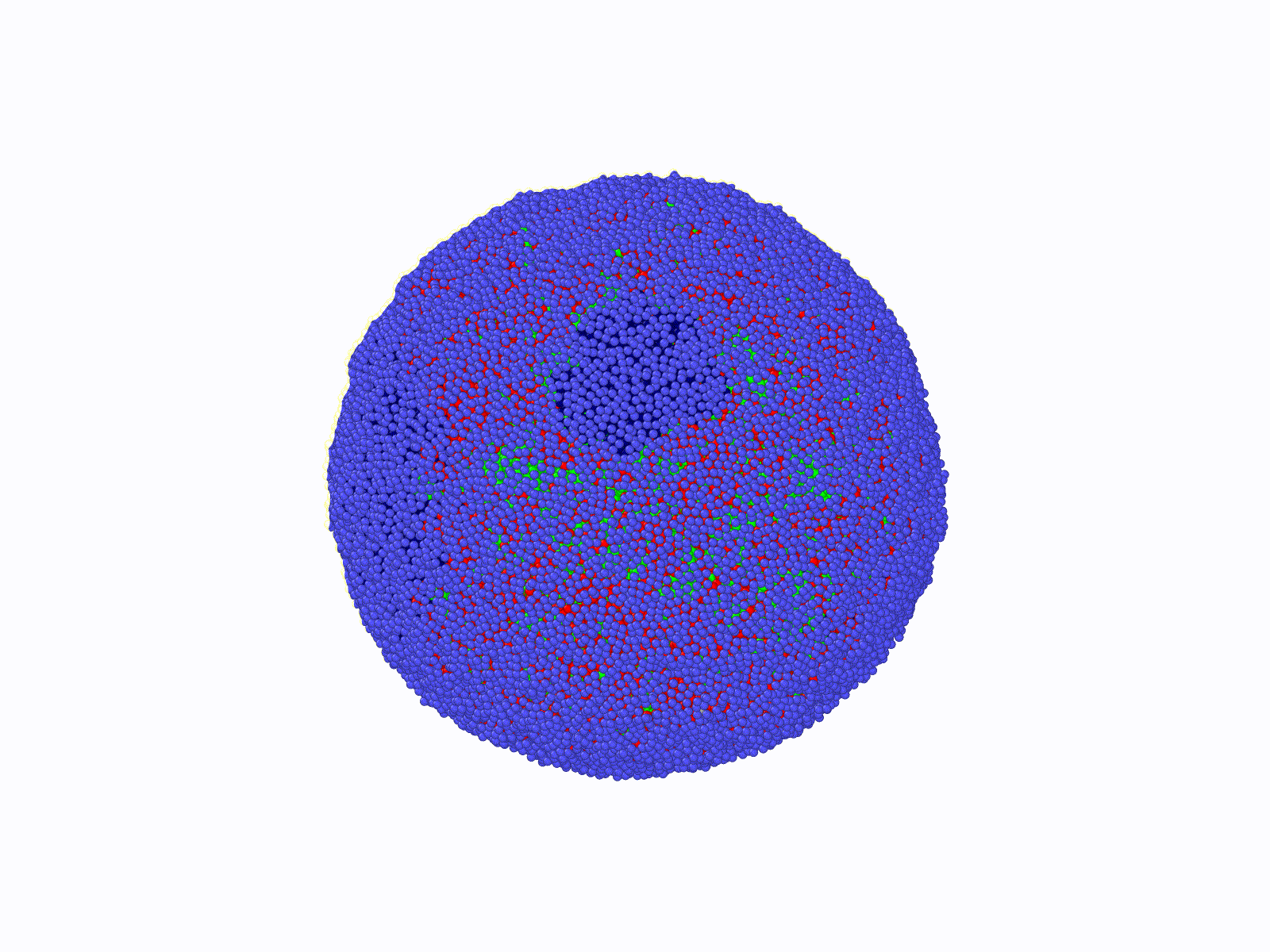
THE SCOPE OF OUR RESEARCH
DNA molecules in our cells are incredibly long polymer chains of nucleic acids that can reach meters in length when appended end to end. These long polymers constituting our genome reside inside a micron-size elastic shell called a cell nucleus. While the nucleus isolates the genome from the rest of the cytoplasm, it also provides a physical template for a highly folded genome. While the nucleus confines a large set of nuclear proteins (i.e., histone) folding DNA polymers into chromatin fibers, it can also maintain a well-defined nuclear chromatin distribution by providing a tethering surface for chromatin at the inner nuclear wall.
Consequently, the chromatin's 3-dimensional (3d) organization within the nucleus and nuclear morphology are highly correlated. Biomolecular experiments have identified abnormal nuclear shapes and impaired nuclear organization of chromatin in many cancer types and genetic diseases, including premature aging in children (Hutchinson-Gilford syndrome). Furthermore, a gradual disruption of nuclear chromatin organization and shape is a hallmark of chronological aging. Notably, nuclei of cancerous cells can become mechanically softer, making it easier for a malignant cell to spread from tissue to tissue. This mechanism suggests that anomalies in the 3d genome organization in a cancerous cell should favor unique nuclear properties compared to a healthy cell's organization. Indeed, detecting alterations in nuclear shape is one of the current (empirical) diagnosis methods for cancer.
All this evidence suggests a strong correlation between the 3d architecture of the genome, nuclear morphology, and genetic function. Understanding such correlations at any level and spatial resolution could generate groundbreaking information. Notably, understanding this complex relationship has been among the priority topics of cell biology and biophysics (e.g., EU Nucleosome Consortium or 4D genome Project of the US National Institute of Health, NIH).
The principal aim of this project is to contribute to the understanding of the relationship between nuclear morphology of mammalian cells and 3d genome architecture in a computational-theoretical framework at a course-grained level. The outcomes of this project can provide roadmaps for future diagnostic systems and shed light on the biomolecular origins of certain genetic disorders. To achieve this, comprehensive, coarse-grained molecular models are designed, and molecular dynamics simulations are conducted. Theoretical models and bio-molecular data analyses will enhance simulation results. To solve this complex and challenging problem, Dr. Aykut Erbaş (PI, molecular bio/physics expert) and Dr. Jaroslaw Paturej (host, theoretical polymer physics, and large-scale molecular simulations expert) have been combining experience and knowledge during the time period of this project supported by NCN (Narodowe Centrum Nauki).

Counters of healthy and cancer cell nuclei labeled by green florescent proteins.
(Retrieved from: Scaffidi et al., 2005; Israeli et al., 2012, Odgren et al., 2010)
(Left) Normal vs Progeria cell nucleus. Right a child with progeria syndrome (Progeria Res. Foundation)
https://www.progeriaresearch.org

